Working with data in MATLAB
Contents
Reading and saving data
Suppose that you have carried out a large computation with MATLAB - one that takes a couple of hours or so to finish. Do you need to run this program every time you want to look at the output? No, you don't - not if you've saved the output to a file.
MATLAB provides many different options for saving data to a file, in many different formats using commands such as
fprintf
or
fwrite
These commands produce data files that can be read into MATLAB or into different programming languages; detailed descriptions of how to work with them can be found using the help command.
The easiest data format to work with in MATLAB -- and the one we'll focus on -- is MATLAB's own native format. Files in this format end with the ".mat" suffix - e.g.
data_out.mat
To save data in the .mat file 'flnm.mat', the command
save flnm.mat
is used. To retrieve data from the .mat file 'flnm.mat', the command
load flnm.mat
is used. Consider the following example, in which we numerically approximate the solution to an ODE
dt = .01; t = 0:dt:10; x(1) = 3; for k=2:length(t) x(k) = x(k-1)+dt*(1-x(k-1)^2); end
We can look at the solution:
plot(t,x)
To save the output to the file data1.mat we type
save data1.mat
We can then clear memory
clear all
If we now try to plot x against t, we'll get an error message - because neither is in memory:
plot(t,x)
Undefined function or variable 't'.
But if we load the data back into memory
load data1.mat
then both of the arrays x and t are back in memory
plot(t,x)
Being able to save to and read from data files is very important when working with programs that take a long time to run (so you don't want to have to do it over and over again), or when working with large datasets.
Working with data
Suppose we've measured the POC concentration in ocean sediments at a number of different depths and we want to estimate the surface sediment flux and refractory POC concentration, knowing that the bioturbation diffusivity is
D = 35 cm^2/kyr
that the remineralization rate coefficient is
k = 1.5x10^-9 s^-1
and that the burial flux rate w is negligible. As we discussed in section 6.5, we can do this using linear regression modelling.
But what are our data? Well, the data file
poc_data.mat
contains measurements of POC (in units mol/m^3) at depths z (measured in m) with the sign convention that z increases with depth. These data are saved in the arrays C and z. This dataset can be downloaded from the course website at
http://web.uvic.ca/~monahana/eos225/poc_data.mat
Download this file to your home directory. Having downloaded the data, let's read them in to MATLAB
load poc_data.mat
and have a look at them
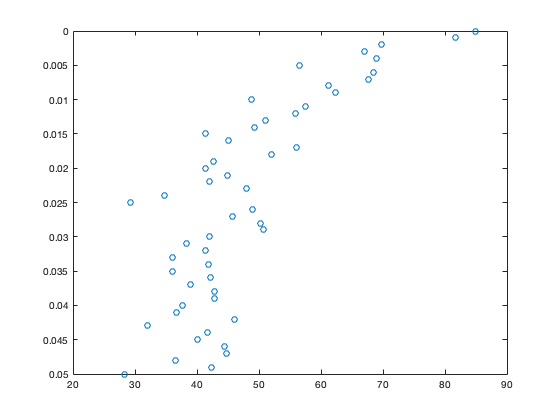
plot(C,z,'o')
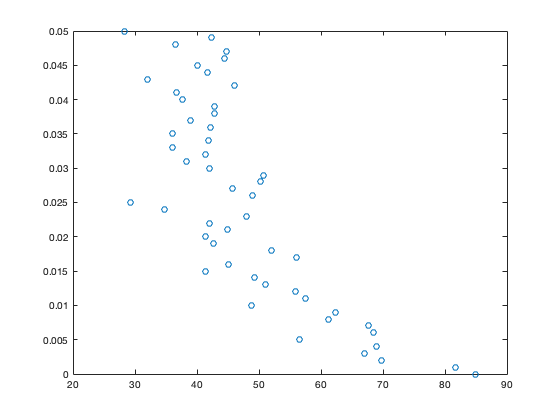
Note that because z increases with depth, deeper values of C are at the top of the plot. We can use the following command to flip the orientation of the plot
set(gca,'ydir','reverse')
The set command can be used to change the plot in all sorts of ways - in this case, to set the y direction ('ydir') from 'normal' to 'reverse' (that is, increasing downward). To get more information, you can type
help set
These data show a general decrease of POC concentration with depth (as expected), but one which is noisy.
From our discussion in class, we know that we can write the POC profile predicted from the steady-state POC budget as the linear regression model
y = ax + b
with
y = C x = (1/sqrt(D*K))*exp(-sqrt(k/D)z) a = Phi b = C_r
To estimate a and b, we'll need to compute some statistics. These computations involve adding up lots of numbers. Such sums are done easily in MATLAB when data is stored as 1xN arrays using the 'sum' command:
sum(M)
which simply adds up all of the elements in the 1xN array M. E.g. with
M = [1 3 8]
M =
1 3 8
then
sum(M)
ans =
12
is just the sum 1+3+8 = 12.
To carry out our statistical analysis, type the following in a text editor and save as poc_stats.m
load poc_data.mat N = length(C); D = 35/((100^2)*(1000*365*24*3600)); %% change D from cm^2/kyr to m^2/s k = 1.5e-9; y = C; x = exp(-sqrt(k/D)*z)/sqrt(k*D); mu_x = sum(x)/N; mu_y = sum(y)/N; sig_x = sqrt(sum((x-mu_x).^2)/(N-1)); sig_y = sqrt(sum((y-mu_y).^2)/(N-1)); rho = sum((x-mu_x).*(y-mu_y))/((N-1)*sig_x*sig_y); a = (sig_y/sig_x)*rho; b = mu_y-rho*sig_y*mu_x/sig_x; Cr = b; Phi = a;
Looking at a scatterplot of x with y:
plot(x,y,'o')
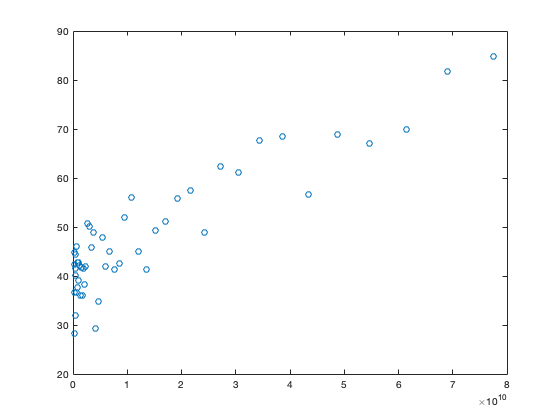
we can compare this to the line predicted by the regression model:
y_reg = a*x+b; hold on plot(x,y_reg,'r') hold off
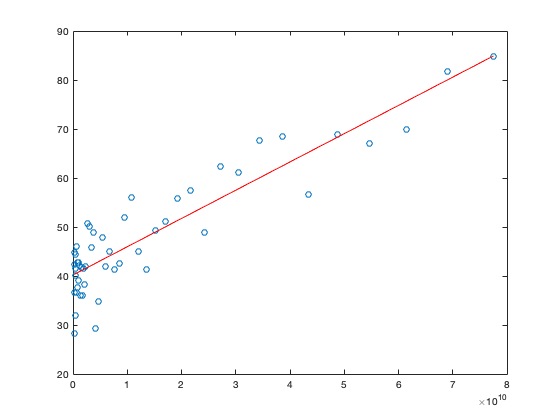
and we see that the regression model does a pretty good job of characterising the scatter of data. The fraction of variance of y "explained" by the regression is
rho^2
ans =
0.8168
and the predicted values of the surface POC flux (in mol m^-2 s^-1) and concentration of refractory POC (in mol m^-3) are
Phi Cr
Phi = 5.7693e-10 Cr = 40.2022
To get Phi in mmol m^-2 day^-1 we have
Phi*1000*24*3600
ans =
0.0498
We can also look at plots not of x against y, but of z against C for both the data and the regression fit. Having computed estimates of Phi and Cr, we have all of the parameters necessary to plot our function C(z).
C_pred = Cr+(Phi/sqrt(k*D))*exp(-sqrt(k/D)*z); plot(C,z*100,'o'),hold on plot(C_pred,z*100,'r'),hold off set(gca,'ydir','reverse') xlabel('C (mol m^{-3})') ylabel('z (cm)')
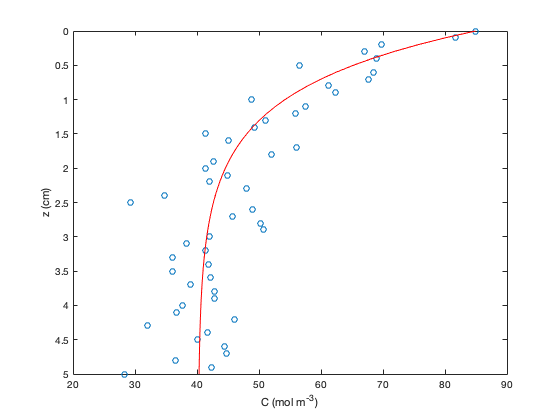
Note that we were able to get a superscript in the x label by typing '^{-3}'. The caret indicates that everything in the curly brackets is in superscript.
Exercise 1
Consider the dataset:
x y -8.6 -3.7 6.6 -4.3 -6.0 -8.4 10.0 7.4 0.13 -1.3 -4.4 0.80 -3.1 -7.1 -5.2 -7.5 -12.3 -8.2 -10.3 -25.2
Assuming the linear regression model y = ax+b, determine least squares estimates of a and b. Plot both the original data and the regression line on the same figure. What fraction of the variance of y is accounted for by this regression?
Exercise 2
Repeat the estimates of Phi and Cr from the dataset poc_data.mat, using data from only the upper 2 cm. How do your estimates change?
Exercise 3
Suppose that for the dataset poc_data.mat you know that Phi = 0.05 mmol m^-2 day^-1 and Cr = 40 mol m^-3. Use linear regression modelling to estimate D and k. Hint: Have a look at Example 6.8.