Bruce
Deagle, Post-Doc
email: bdeagle@uvic.ca
The focus of my research has been using information in DNA
to address ecological and evolutionary questions. My PhD thesis looked at
methods to study the diet of large marine predators by analysing prey DNA in
their faeces. In August 2008 I switched gears and started a stickleback
evolutionary genetics project in Tom Reimchen’s laboratory, we will be
working in close collaboration with David
Kingsley’s group at
Postdoctoral work: SNP variation in Haida Gwaii
threespine stickleback

Studies of Haida Gwaii stickleback
over the last 35 years have documented a remarkable level of morphological
variation among freshwater populations. These populations have been the focus
of numerous evolutionary investigations and provide a textbook example of
adaptive morphological evolution (see Reimchen publications).
In this postdoctoral project we will obtain a genomic perspective of the
genetic structure and history of these stickleback using a genome-wide SNP
(single nucleotide polymorphisms) genotyping system developed at Stanford.
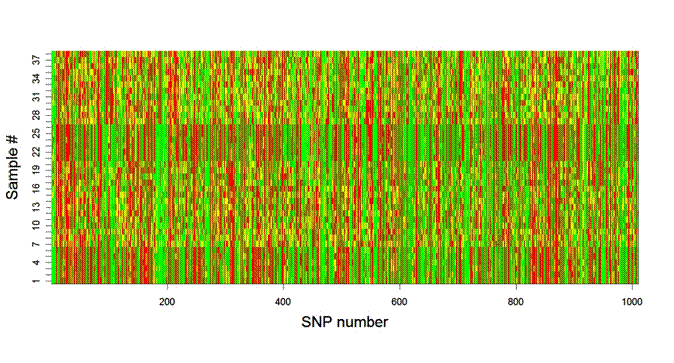
Figure 1: Image plot of preliminary SNP
data (1010 loci) from 26 Haida Gwaii and 12 marine stickleback.
Red
or green = homozygous, yellow = heterozygous.
PhD work at the University of Tasmania:
DNA-based methods to study diet
Studying animal diet is a difficult undertaking especially
in the marine environment where it is rarely possible to observe feeding
activity. Some prey remains survive digestion and morphological analysis of
remains in faecal material can provide an indication of what predators are
eating. Analysis of stomach contents is also possible, but this involves
invasive or lethal sampling. In my PhD I looked at the potential for use of
DNA-based identification methods in dietary studies of marine predators. When I
started it was clear that prey DNA was present in animal faeces but how useful
it would be for dietary studies was an open question. So, in collaboration with
researchers from the Marine Mammal
Unit at UBC, I started by looking at samples collected from captive Steller
sea lions to address some basic questions:
·
 Can prey DNA be reliably detected in the soft matrix of faecal
samples?
Can prey DNA be reliably detected in the soft matrix of faecal
samples?
·
Can DNA from prey items fed as a
small proportion of the diet be detected?
·
Are the relative amounts of DNA
recovered from prey species proportional to their mass in the diet?
·
What is the quality of the prey
DNA recovered?
These questions
are the focus of three papers from my thesis:
Deagle
BE, Tollit DJ, Jarman SN, Hindell MA, Trites AW, Gales NJ (2005) Molecular
scatology as a tool to study diet: analysis of prey DNA in scats from captive
Steller sea lions. Molecular Ecology 14: 1831–1842. (pdf)
Deagle
BE, Eveson JP, Jarman SN (2006) Quantification of damage in DNA recovered from
highly degraded samples — a case study on DNA in faeces. Frontiers in
Zoology e3:11. (pdf)
Deagle
BE, Tollit DJ (2007) Quantitative analysis of prey DNA in pinniped faeces:
potential to estimate diet composition? Conservation Genetics 8: 743–747.
(pdf)
The methods
were further evaluated in a field-based study looking at the diet of macaroni
penguins at Heard Island and most
recently I have been involved in a DNA-based diet study of Australian fur seals
in Bass Strait, southern
Deagle
BE, Gales NJ, Evans K, Jarman SN, Robinson S, Trebilco R, Hindell MA (2007)
Studying seabird diet through genetic analysis of faeces: a case study on
macaroni penguins (Eudyptes chrysolophus).
PLoS ONE 2: e831. (pdf)
Deagle
BE, Jarman SN,


For my other papers, please see publication list.
Funding:











